Data Science
Guide
Data Science—also called Data Analysis—is a set of operations that most often occurs after birth and death information have been collected through standard SRS operations. Key steps and processes for data science are enumerated below, and many candidate tools and software systems exist to support these steps.
Data Quality Checks
Data quality checks should occur at all stages of data collection. While the forms and data collection systems should include data validation checks as the questionnaires are administered, the SRS team member in charge of data quality and review should complete additional checks. In many countries, this individual will be a Data Manager, either at a regional or national level. The checks can be broken into three categories by the three types of SRS events (i.e., pregnancies, pregnancy outcomes, and deaths). This section presumes that Verbal Autopsy is the method used to ascertain Cause of Death; however, other methods (such as Minimally Invasive Tissue Sampling [MITS]) can be used. Data quality checks should still occur in this case, though they may be different than what is described here.
Similar to Operational Reporting, defining the data quality and assurance checks are not the responsibility of the IT team; rather, the SRS epidemiologists and program staff should define the checks based on global guidance and best practice and convey those checks to the IT team. However, there exist some commonly-used data quality checks, and they are presented in this section to provide awareness and context to the IT teams about what they might need to track in the SRS systems.
Pregnancy & Pregnancy Outcomes
For pregnancy and pregnancy outcome data, data quality checks should ensure:
- All pregnancy events have at least one associated pregnancy outcome
- A single fetus does not have conflicting outcomes
- A death event is recorded for any relevant pregnancy outcomes (e.g., stillbirths)
Tech Tip: Multiple Births
In the event of a multiple birth, a single pregnancy event could result in multiple pregnancy outcomes. While the forms should support this result, data quality checks should be written to acknowledge that the number of pregnancy outcomes per pregnancy event may not be a 1:1 mapping.
Verbal Autopsy
If using the WHO Standard Verbal Autopsy Instrument, the XLSForm specification distributed by WHO includes built in quality checks that run as the questionnaire is administered. However, similarly to births, the Data Manager should run additional checks including:
- De-duplication: Identify duplicate records based on Study IDs and investigate completeness of each record to determine which record to drop or merge.
- Consent: Ensure that the individual gave consent to VA interview. This can be checked by looking for
Id10013 = "YES"and, in the event of pausing and re-starting the interview, ensure this condition continues to hold. - Completion time: Flag individual interviews with completion time <15 minutes and >90 minutes for further investigation. A standard VA interview should take place within those bounds. Additionally, aggregate these statistics up to the interviewer to identify any interviewers who consistently conduct interviews that do not meet these criteria, which could indicate a need for additional training.
Cause of Death Assignment
Computer Coded VA (CCVA)
Multiple computer algorithms exist for assigning COD programmatically. Most, if not all, of these algorithms are useful to determine causes of death in aggregate across an entire population; however, they should not be used to investigate COD at an individual level.
Tech Tip: CCVA Algorithm & WHO Instrument Version
Certain CCVA algorithms are only available for specific versions of the WHO standard VA instrument. Specifically, the 2022 standard instrument can only be used with unvalidated CCVA algorithms such as InterVA. These unvalidated algorithms may be good enough for your country’s use case, but that is a decision that should be made in consultation with the entire SRS team.
If you choose to use the 2022 standard questionnaire, then make sure to either transform the questionnaire back to the previous version to use the validated algorithms; or, ensure that all data are reported with an acknowledgment that unvalidated algorithms were used to determine COD.
Physician Coded VA (PCVA)
Physician Coded VA assigns COD by asking a panel of physicians to review the VA results and determine the most accurate cause of death. While PCVA often produces more accurate results than CCVA, it is typically not feasible to conduct PCVA at scale with the size of a SRS program. Therefore, PCVA is recommended for validation use or other targeted use cases, as opposed to general adoption for SRS processes.
Epidemiological Reporting and Data Use
Data analysis platforms also need to be able to support the use of SRS data for epidemiological purposes and national reporting. While the global SRS, CRVS, and VA community have developed ample guidance on data analysis and interpretation—some of which is linked below—this analysis must be developed based on the needs of the data users in your country. Conduct stakeholder engagement and listening sessions to understand the types of reports that the epidemiologists and statisticians need to create from SRS data. Those are the reports and calculations that should be built directly into your system.
Tools
Data Quality
-
Data Review Log Form (docx) A summary of how to utilize a data review log form to identify possible data collection errors.
-
Data Review Feedback Form (docx) A summary of how to utilize a data review feedback to provide detailed analyses of CSA activity.
Data Processing & Analysis
-
Data Processing Steps (docx) A summary of data management processes for SRS data.
-
Basic Steps for Mortality Rates (docx) A guide on how to calculate mortality rates and ratios.
-
Basic Steps for Cause of Death Analysis (docx) A guide on how to implement Computer-Coded Verbal Autopsy (CCVA) algorithms to determine causes of death.
-
Guidelines for interpreting verbal autopsy data (pdf) A document developed by Bloomberg Philanthropies Data for Health Initiative outlining five steps for users of VA to follow to help them interpret and present their VA data.
If Social Autopsy is being implemented as part of SRS:
-
Social Autopsy Analysis Guide for Children Under 5 and 5-17 (docx) A guide on methods for analyzing child social autopsy data.
-
Social Autopsy Analysis Guide for Adults 18-50+ (docx) A guide on methods for analyzing adult social autopsy data.
Software
CCVA algorithms
Multiple algorithms to perform CCVA exist, and a specific algorithm or suite of algorithms is not recommended for SRS operations. The following table highlights available options, providing a description of each algorithm and a link to the source package. Most, but not all, of these CCVA algorithms are available as R libraries. The goal of each algorithm is the same: programmatically and algorithmicly assign cause of death to a verbal autopsy. The algorithms differ by method and—sometimes—by other items such as the possible range of causes.
In alphabetical order. Note that this list may not be exhaustive.
| Algorithm | Description |
|---|---|
| Expert Algorithm Verbal Autopsy (EAVA) | Expert Algorithm Verbal Autopsy assigns causes of death to 2016 WHO Verbal Autopsy Questionnaire data. This algorithm uses the presence and absence of signs and symptoms reported in the Verbal Autopsy interview to diagnose common causes of death. A deterministic algorithm assigns a single cause of death to each Verbal Autopsy interview record using a hierarchy of all common causes for neonates or children 1 to 59 months of age. Integrates with the openVA wrapper library available in R that offers multiple CCVA algorithms in one, but not maintained by the openVA team. |
| InterVA | An R package replicating InterVA software for coding cause of death from verbal autopsies collected using the 2012 and 2016 WHO VA instrument. It also provides simple graphical representation of individual and population level statistics. Maintained alongside the openVA wrapper library available in R that offers multiple CCVA algorithms in one. |
| InsilicoVA | An R package for using the InSilicoVA algorithm for coding cause of death from verbal autopsies collected using the 2012 and 2016 WHO VA instruments. It also provides simple graphical representation of individual and population level statistics. Maintained alongside the openVA wrapper library available in R that offers multiple CCVA algorithms in one. |
| Tariff 2.0 / SmartVA | SmartVA-Analyze is an application that implements the Tariff 2.0 Method for computer certification of verbal autopsies. It takes verbal VA data as input and produces cause of death estimates at the individual and population levels. The SmartVA cause of death assignment system was designed and validated by the Institute for Health Metrics and Evaluation (IMHE) with the Population Health Metrics Research Consortium (PHMRC) Gold Standard VA database collected as part of the PHMRC Gold Standard VA Validation Study. An R package exists to implement the algorithm; however, it was not developed by the original authors. For the most accurate and up-to-date Tariff implementation, you should use the original software developed by IHME, linked at left. |
Candidate CCVA algorithms often expect data in different input formats. Some of the algorithm packages come with data transformation tools such as the odk2EAVA that comes with the EAVA algorithm package. Other data transformations can be performed with the pyCrossVA transformation library available in Python.
Lastly, the vacalibration R package provides a computerized approach to calibration of CCVA algorithms by leveraging gold standard causes of death and misclassification matrix framework (link). This is not something that will likely be a part of daily SRS operations; however, it could be useful for SRS program setup or evaluation.
Integrated processing & data visualization
VA Explorer (VAE)
VA Explorer (VAE), originally developed under a CDC Foundation and Bloomberg Data for Health Initiative to facilitate administration and analysis of VA programs, was in use in Zambia at the Ministry of Health from 2020-2025. It is an open source piece of software that integrates with CCVA algorithms and data processing, providing a graphical interface for VA reporting and analysis. For more on how Zambia has adapted this software to SRS use, see the Zambia story below.
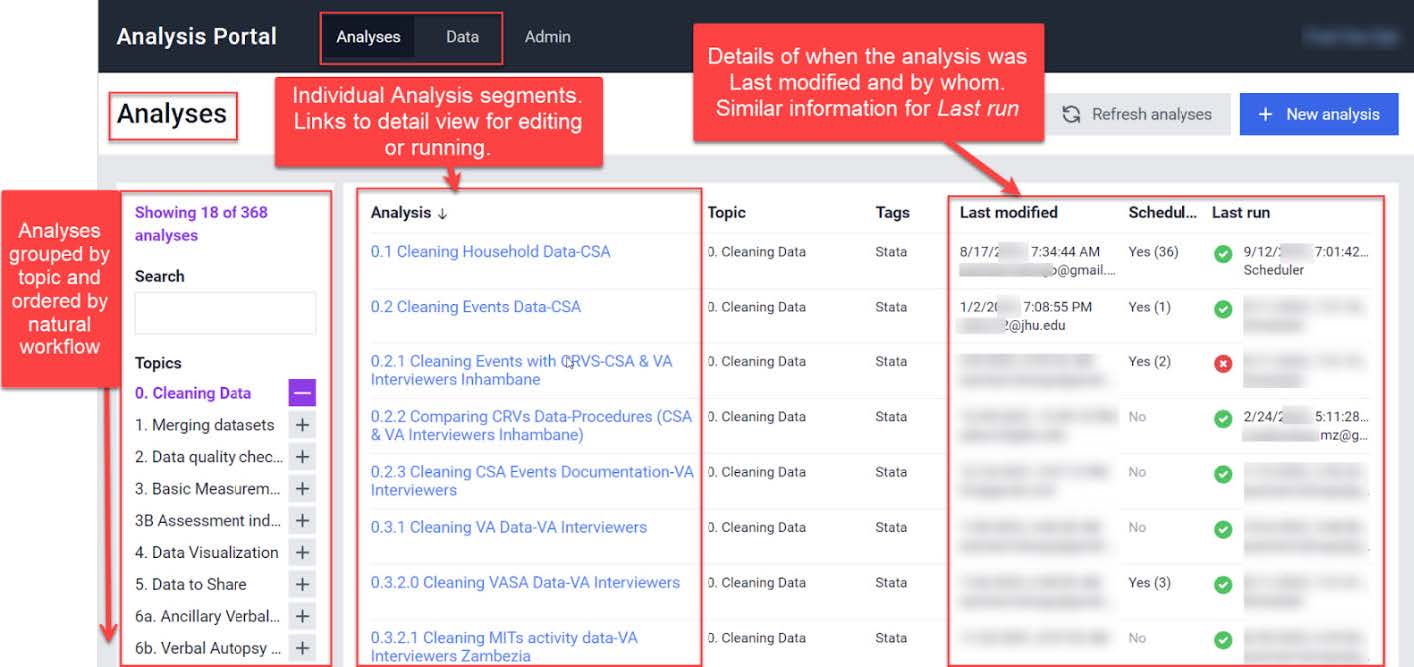
COMSA/SISCOVE Analysis Portal
The COMSA/SISCOVE Analysis Portal, discussed more below in the Mozambique story, is a tool that can be deployed using Docker to facilitate collaborative analysis of SRS data housed in ODK.
Tech Tip: Integrated Analysis
For an integrated analysis system for death data, consider using VA Explorer in your country. While the current versions of VA Explorer do not support additional SRS data such as pregnancy and pregnancy outcomes or social autopsy, other countries are currently adapting the system for such uses. This page will be updated when appropriate.
Stories
Zambia
ZNPHI VA Explorer Development
When implementing SRS, Zambia had existing systems in use in country for Verbal Autopsy administration that they chose to adapt for SRS purposes. To determine how to adapt those systems, they held a stakeholder workshop attended jointly by IT team members and SRS program staff, epidemiologists, and statisticians. This workshop served to lay a common foundation and to quickly build consensus of the role of the IT systems. Activities conducted in this workshop that were particularly helpful were a Premortem at the beginning of Day 1 to establish a shared understanding of success and spending most of Day 2 collaboratively defining Personas, a generic descriptive model of a user who will interact with the SRS IT tools.
A set of workshop slides specific to SRS that were used during this ZNPHI IT stakeholder workshop can be found here.
Following this workshop, the IT team had sequential meetings to create a list of software requirements for the IT system, conducted a gap analysis to determine what needed to change in existing systems, and then established a group of software developers to make those changes. Once the requirements were defined and the IT team understood what needed to be built, initial software development was complete in about 3 months. However, the requirement definition process took about 9 months before a single line of code was written, while being originally estimated as something that would not take significant amounts of time. Zambia’s experience highlights the critical role of stakeholder engagement, and underscores that building a shared understanding to develop software takes longer than you think.
Mozambique
COMSA/SIS-COVE Analysis Portal
As part of the Mozambique SRS program, the SRS team, alongside Johns Hopkins University, created a collaborative monitoring and analysis server (Analysis Portal or just Portal) was created to host analytic datasets and codes, and to allow data analysis collaboration for the SRS project team. The Portal uses a Linux Cloud Server and a custom web platform including React, Docker, Linux, Stata, R software. The master data sits in the underlying SQL database (as discussed in Infrastructure & Databases), and the Portal site can access a copy of the master data such that any changes to the data on Portal do not affect the underlying data.
The Analysis Portal has a data pool with two types of data. First, there is the data collection data from the ODK platform. This data is automatically copied from the data collection server to the Portal every night in the form of text-based files. Second, the Portal has supplemental, or reference data, that can be uploaded manually.
The Analysis Portal has a page dedicated to analysis where scripts can be sequentially and automatically run on the data. In general, the cleaning, merging, and quality check scripts are scheduled to run after the daily data synchronization. This provides daily updates for teams monitoring the data and allows real-time analytic collaboration across multiple continents.
The following screenshot provides a view of the Analysis portal, illustrating how scripts in multiple languages can be edited and run collaboratively.
| Last updated |
|---|
| 9 January 2026 |
| Portions of this page are © 2026 The MITRE Corporation. All rights reserved. Approved for Public Release #25-2779. Distribution Unlimited. The source of this information is the Technical Assistance for Sample Registration Systems (SRS) Planning Grants, a joint project of the CDC Foundation and Swiss Tropical and Public Health Institute through the Gates Foundation SRS Grant. |